Researchers solve some of the drawbacks related to DNA-based information storage by using synthetic macromolecules as an alternative for data encoding.
In this age of information, keeping data safe is of the utmost importance. Our desire to conceal information from unwanted eyes has been documented throughout history, from invisible inks — which were used for hundreds, if not thousands, of years — to the advanced cryptography used today to protect digital data.
But now science has taken this to the next level with the emerging field of molecular cryptography, in which information is hidden within molecular or biomolecular encryption systems. In addition, as we generate more and more data, we will soon exceed our capabilities to store it using silicon-based chips, and molecule-based storage systems have the potential to expand our storage capacity.
Given its ability to store, process, and transmit vast amounts of information, DNA has been an obvious front-runner. But while biological information storage has already been demonstrated using DNA, there are drawbacks such as a restricted number of natural building blocks, limited re-writability, susceptibility to degradation, and cost.
Therefore, in a recent paper published in Advanced Science, researchers from Ghent University have set out to solve some of the drawbacks related to DNA-based information storage by using synthetic macromolecules as an alternative for data encoding.
“Although many protocols have been developed over recent years to store information on non‐natural, sequence‐defined macromolecules, data encryption still remains an important aspect in future molecular data management, as individuals and enterprises seek more sophisticated and fraud‐resistant technologies,” stated the authors in their study.
To do this, the team created a procedure to reversibly write a molecular PIN code using a series of molecular building blocks that could be assembled into a variety of combinations using a multi-component reaction — called the Passerini 3-Component Reaction — used broadly in synthetic chemistry. By changing one of the reagents the team was able to alter the nature of the linkage between monomers to make the sequence’s backbone a thermally reversible bond — this means that when the PIN sequence gets heated it will readily break apart.
Specific molecular components in the reaction were designated the numbers one to four (numbers for the PIN) and were linked with molecular markers or fingerprints that could be easily identifiable using mass analysis. These molecular markers are important for read-out as they each bear a distinct isotopic pattern, which can be used to mark the digit’s position within the sequence during mass analysis.
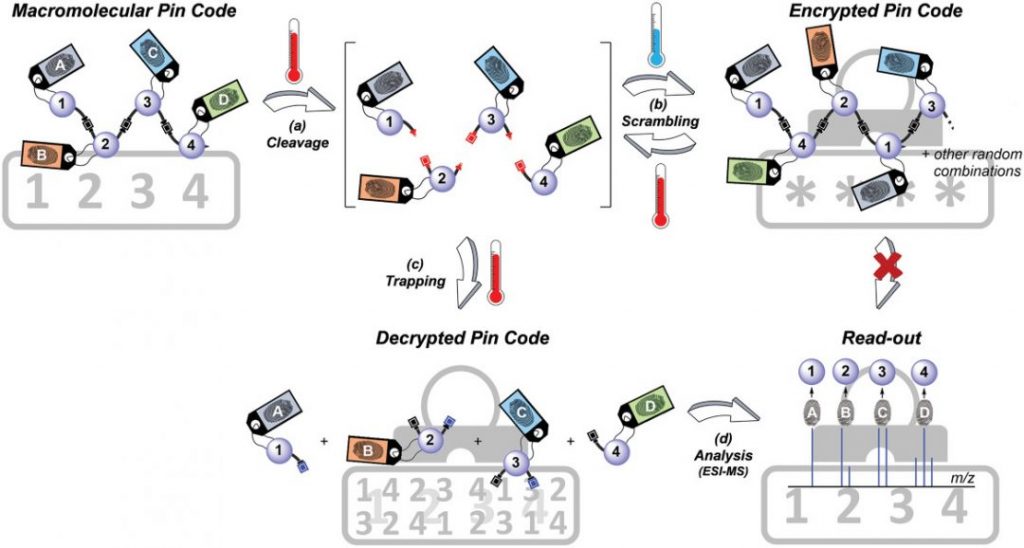
Through a two step, iterative process, the team was able to chemically write the numeric PIN code, 1-2-3-4, with the number’s positions identifiable as a result of fingerprints designated A-B-C-D. Once created, the code was then heated to break and scramble it, as when the mixture cools it results in the recombination of PIN numbers in a completely random order.
To retrieve the encrypted information, the scrambled sequence was heated in the presence of a molecular trap — a molecule that forms a more favorable bond with the now separated monomer units than they do with each other. The now, isolated monomers were then easily identified using mass analysis and the original code identified based on the monomer’s attached fingerprint.
“The use of four different isocyanides [the molecular PIN numbers] allows for 44 (256) possible combinations, which translates to a small storage capacity of 8 bits. Our strategy can, however, readily be expanded to a wide range of other isocyanide compounds, thereby significantly extending the amount of codable digits to increase the amount of possible permutations to n4,” said the authors. “The implementation of a reliable, dynamic covalent chemistry within sequence‐defined macromolecules could be considered an important advancement in the development of the next generation of encryption and decryption technologies.”
Research article found at J.O. Holloway, et al. Advanced Science, 2020, doi.org/10.1002/advs.201903698

