New imaging technique sheds light on the mysterious life cycle of giant viruses.
In 1992, the first known giant virus, Acanthamoeba polyphaga mimivirus (APMV), was discovered by microbiologist Timothy Robotham in a power-plant cooling tower in Bradford, England. At the time, scientists were investigating the cause of a local pneumonia outbreak, and due to its size, APMV, named mimivirus — short for mimicking microbe — was incorrectly identified as new type of pneumonia-causing bacteria. It wasn’t until a decade later that scientists realized they had not identified a new type of bacteria, but a new class of virus.
In subsequent years, new examples of giant viruses have been discovered around the world including an ancient, 30,000-year-old giant virus named Pithosvirus, which was thawed from permafrost in Siberia and still active and able to infect host cells today.
These viruses are so large they can be viewed under a microscope (hence the initial classification of mimivirus as a bacterium) and have certainly changed our perception of these microbes. However, it’s not only their size that has puzzled and captivated scientists, but their incredibly complex genomes, which dwarf their smaller counterparts. Mimivirus, which is a giant among giants at 750nm in diameter (exceeding the size of several well-characterized bacteria), contains approximately 1.2 million base pairs and codes for 1,018 genes; the HIV virus, on the other hand, has roughly 12 genes. What’s also so intriguing is the fact that the genome of giant viruses encodes for their own translational machinery, where smaller viruses hijack that of their host cell. This was previously thought to be characteristic of cellular organisms, but has instead turned this idea on its head, triggering debate about the origin of these gigantic microbes.
While research in this area began almost twenty years ago, little is still known about the life cycle of giant viruses due to their gargantuan size and complexity. When we think of a viral infection, we imagine a small, covert agent gaining entry to the cell, overriding that cells machinery to replicate to the point where the cell bursts, releasing new viruses to continue the cycle of infection.
What then are the conditions and mechanisms that these enormous, bumbling microbes use to infect host cells? It is known that giant viruses, such as mimivirus, gain entry into phagocytic cells (such as amoebae) when these scavenger cells engulf the virus along with other food. It enters the amoebae’s cytoplasm and begins releasing proteins to begin its takeover, but how exactly this happens has remained unclear.
A new study recently published in the journal Cell sought to answer some of these questions using a new, cutting-edge imaging technique to study giant viruses and how they infect host cells.
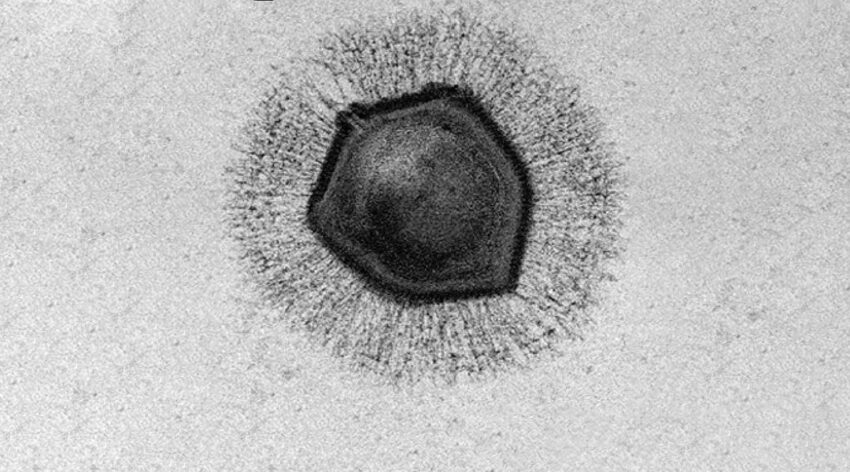
According to the team led by Kristin Parent, associate professor of biochemistry and molecular biology at Michigan State University, giant viruses have large, durable outer shells, called capsids, that help them withstand harsh environments — such a being frozen for millennia, just say. In the current study, Parent and her team were able to uncover some of the mystery surrounding giant virus infection mechanisms by determining the biological conditions that promote genome release during infection.
All of the giant viruses used in the study have icosahedral outer shells shaped like a twenty-sided die. At the top of one of the shell’s vertices lies a starfish-shaped seal called the “stargate”, which opens during infection to release the viral genome. How and when this happens has long been a mystery. According to Parent, this is because giant viruses have been difficult to image due to their size, and “previous studies relied on finding the ‘one-in-a-million’ virus in the correct state of infection”.
To solve this issue, the team used cryoelectron microscopy (cryo-EM), cryoelectron tomography (cryo-ET), and scanning electron microscopy (SEM) to observe the viruses under an array of harsh chemical and environmental treatments designed to simulate conditions a virus might experience during the infection process.
During experimentation it was found that three environmental conditions — low pH, high temperature, and high salt — successfully induced “stargate opening”, and a different stage of infection. This allowed the researchers to create a model to reliably mimic the virus’ behavior, which according to Parent, will simplify future study of its life cycle.
Another key finding of the study was that the starfish seal above the stargate portal slowly unzips while remaining attached to the capsid, resulting in a slow release of the virus’ genetic material. “We discovered that the starfish seal above the stargate portal slowly unzips while remaining attached to the capsid rather than simply releasing all at once,” Parent said. “Our description of a new giant virus genome release strategy signifies another paradigm shift in our understanding of virology.”
The team’s approach also allowed them to study the proteins being release during the initial infection stage. “The results of this study help to assign putative — or assumed — roles to many proteins with previously unknown functions, highlighting the power of this new model,” Parent said. “We identified key proteins released during the initial stages of infection responsible for helping mediate the process and complete the viral takeover.”
“The exact functions of many of these proteins and how they orchestrate giant virus infection are prime candidates for future study,” Parent continued. “Many of the proteins we identified matched proteins that one would expect to be released during the initial stages of viral infections. This greatly supports our hypothesis that the in vitro stages generated in this study are reflective of those that occur in vivo.”
Quotes adapted from original press release provided by Michigan State University