Researchers use CRISPR-Cas12a as a DNA shredder to clean up and mobilize parts of DNA-origami structures.
DNA origami is the folding of DNA strands into precise shapes on the nanoscale. The technique uses hundreds of short DNA oligonucleotides, called staple strands, to fold a long DNA strand into the desired nanoscale architecture. In recent decades, the budding field has shown enormous potential in drug delivery, biosensing, and biocomputing due to the versatility and complexity that is achievable in DNA-based engineered structures with programmable shapes, patterns, and motions.
However, building a DNA structure from the bottom up using a single, continuous strand does come with some limitations, such as the ability to quickly swap-out different components, make modifications once the structure is built, or create easily create a functioning nanomachine with moving parts.
To address this, a team of researchers from Yale University propose that there needs to be a method to break scaffolds of pre-folded DNA structures to make specific changes while also preserving its overall integrity. In a study recently published in Angewandte Chemie, the team proposes a means of reconfiguring DNA-origami structures using the CRISPR-Cas12a system.
“DNA-origami is a powerful way to engineer nanodevices. However, generating controlled motions of DNA-origami structures often requires complex, multi-step synthesis. This is because a DNA-origami structures have a continuous DNA scaffold strand going through all its domains to hold it intact,” says Chenxiang Lin, professor at Yale University and one of the study’s lead authors. “For assembly and integrity, this is great. However, to introduce dynamic aspects to the origami, the ‘intactness’ becomes a problem i.e., the scaffold strand constrains motion.”
Clustered regularly interspaced short palindromic repeats (CRISPR) proteins were originally components of bacterial immune systems, and are RNA-guided enzymes that bind and cleave DNA. CRISPR-Cas9 was the first to be isolated, though the CRISPR-Cas12a protein used in the current study has some distinguishing features that have expanded the capabilities of these genome-editing tools; CRISPR-Cas12a has found numerous applications in genome manipulation and DNA sensing, among many others.
In the current study, the researchers set out to demonstrate that single strand DNA-specific nuclease CRISPR-Cas12a could be used as a versatile tool to modify the structure of DNA architectures through the selective “trimming” of DNA scaffold or staple strands.
“Using a car manufacturing analogy, it would be as though an entire car was 3D-printed in one step with all its wheels, doors, and gears physically tethered to each other, as well as with a bulk of unused material remaining stuck to the car,” says Lin. “Such a vehicle would not function as the interconnecting tethers immobilized its wheels and gears, and the leftover material might also hinder movement. In our work, CRISPR-Cas12a functions as a cutter to efficiently remove only the tethers between the one-step produced machine components and any leftover material, thereby enabling the machine to perform its dynamic function.”
The team began by demonstrating CRISPR-Cas12’s ability to selectively cleave an unfolded DNA strand from a DNA-origami semicircle. Handles incorporated into the DNA semi-circles for chemical modifications, such as the incorporation of fluorophores, lipids, or other proteins, were pre-hybridized with an “anti-handle” to prevent their cleavage by CRISPR-Cas12a.
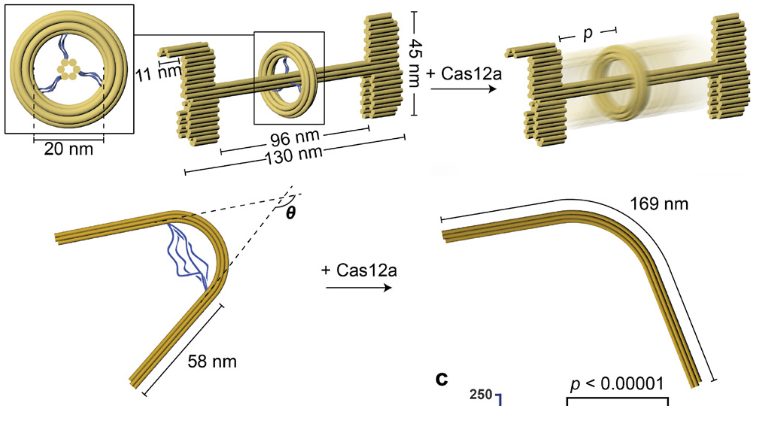
The next step involved attempting to release structural modules from a multi-component DNA assembly through selective cleavage of tethered scaffold strands. As proof of concept, the team designed an origami structure with a rod threaded through three circles, each tethered by three pairs of single stranded DNA. By selectively protecting certain single stranded DNA tethers through pre-hybridization, CRISPR-Cas12as was able to selectively release a desired circle from the rod.
The implications of this are far reaching as this method could be used to trigger conformational changes in a DNA-origami structure, potentially enabling greater engineering control without the complexities of multicomponent, stepwise self-assembly.
To demonstrate this, the group created a nanoscale pseudo-rotaxane, a mechanically interlocked molecular architecture that operates as an artificial molecular machine for molecular switches. Past attempts to build DNA-origami rotaxanes required multi-step assembly of various components made from different scaffold strands and a strand-displacement-mediated release of the macrocycle. In the current study, components of the rotaxane (the ring, stoppers, and rod) were created from the same single strand of DNA in one pot, with selective liberation of the ring demonstrated with the enzyme.
The team also extended this toward designomg a prestressed elastic beam structure that upon Cas12a-processing, releases its elastic potential energy and changes conformation. “We envision incorporating the elastic beam’s prestressed, switchable design into DNA ‘nanosprings’,” said the authors in their study.
“As with any method development in DNA nanotechnology, we want to achieve more precise spatiotemporal control of our Cas12a-controlled devices in the future,” adds Lin. “But perhaps more importantly, we are eager to apply this method to build DNA-based devices similar to protein machineries in cells, [such as] membrane remodeling protein complexes. A long-term goal is to deploy DNA devices to biological systems, where Cas12a activates them (e.g. drug carriers) in response to biochemical cues.”
Cas12a with switchable activities has promising applications in vivo, such as targeted activation of functional nanodevices in response to nucleic acid inputs in different cellular states. “Our work provides a straightforward yet powerful way to build and operate dynamic nanodevices,” says Lin. “Given the ability of CRISPR-Cas12a to integrate nucleic acid inputs, this method could facilitate targeted device activation in cells.”

